How DNA Is Made Available for Replication
Proteins are the central building material of our bodies. As a fundamental component of the cell as well as of DNA and its genes, they make us who we are. Scientists who study the complex interactions of proteins often use high-performance computing (HPC) resources to reveal what otherwise would remain hidden in experiments alone. Recently, an international team of researchers led by the research group of Ben Schuler, professor of molecular biophysics at the University of Zurich, succeeded for the first time in using HPC to decipher the process that activates the DNA in the cell nucleus at the molecular level.
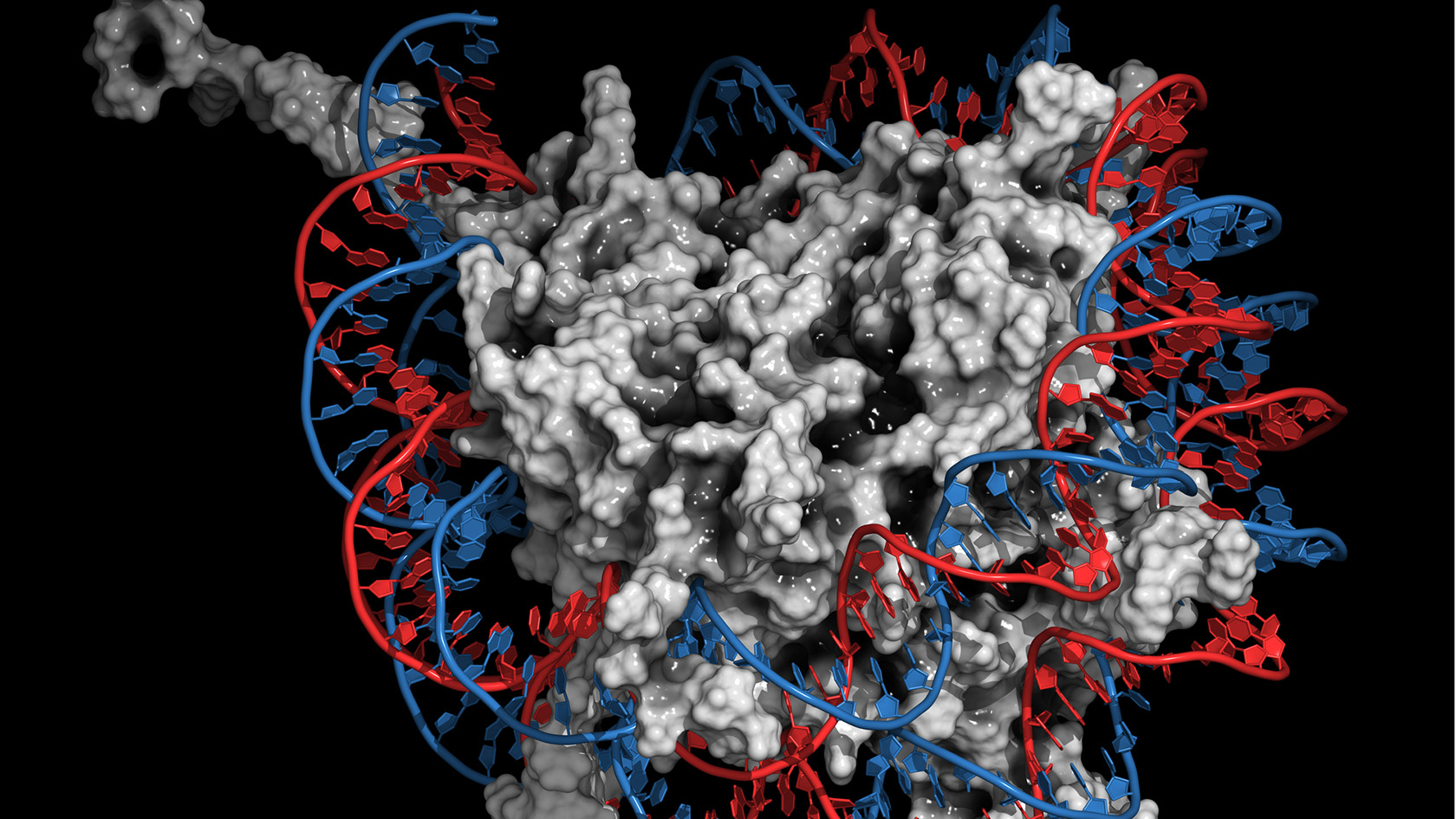
For this process it is essential that the packaging of the DNA in the so-called nucleosome is loosened. The nucleosome is a unit of DNA chains that wrap around histones, which are proteins that are important for packaging. Nucleosomes are the central components of chromatin, in which the approximately two-metre-long chains of DNA in the cell nucleus are compressed into a tiny space of about one 1000 cubic micrometres. In this packaging process, a specific, strongly positively charged histone (H1) helps to seal and compact the strongly negatively charged DNA. In order to loosen this tight packaging, for example, to replicate or repair the DNA, this histone H1 must be detached.
Charged proteins as major players
Although it was previously unclear how the strong binding between histone H1 and DNA could be broken within a useful period of time, the researchers succeeded in deciphering the process with a combination of experiments and simulations on the "Piz Daint" supercomputer at CSCS. According to the results, the unstructured regions of histone H1 and a protein called prothymosin alpha play a central role in the process. In contrast to the positively charged H1, prothymosin alpha has a high negative charge.
Based on the experiments carried out in the laboratory, the researchers already suspected that, in addition to H1, prothymosin alpha is also crucial. It was observed interacting with the H1 via its strong charge and accelerating the detachment process — but it was only the simulations that made the details visible. The researchers recently published the results of their study in the journal Nature Chemistry.
Unique observations
The researchers used the results of the simulations to visualise how prothymosin alpha and H1 bind to each other via their unstructured protein regions. The visualisation shows how prothymosin alpha, H1, and the DNA form a short-lived complex. "No one has observed this in this form before," says Schuler. "These unstructured proteins seem to enable an interaction that is very difficult to explain via traditional structural biology." Specifically, the connection between prothymosin alpha and H1 weakens the interaction between positively charged H1 and negatively charged DNA, so that prothymosin alpha gets the upper hand, so to speak. It detaches the histone from the DNA and transports it away, leaving the DNA ready for replication or repair.
Schuler’s research group specialises in so-called single-molecule spectroscopy, which proved decisive for this discovery in combination with simulations. Until about ten years ago, the focus of research was on the well-defined, folded three-dimensional structure of proteins. "In contrast, the unstructured segments, which occur in about two-thirds of the proteins in our bodies, received little attention in traditional structural biology," says Schuler.
Only the tip of the iceberg
Schuler is convinced that the present findings are only the tip of the iceberg when it comes to researching the role of unstructured proteins in cellular processes. According to the biophysicist, it is already becoming apparent that there are further examples in which unstructured charged proteins in the cell enable further exchange reactions. Schuler and his team are already researching similar mechanisms that play a role in DNA repair.